Click button "Structure Analysis" in PGAP-X Lite Launcher, you will get the "Structure Analysis" interface like Figure 4.0.1. Including three functional buttons:
| You are here: PGAP-X Home > Manual > Get Genome Alignment Data and Visualization Result | ||||
Click button "Structure Analysis" in PGAP-X Lite Launcher, you will get the "Structure Analysis" interface like Figure 4.0.1. Including three functional buttons:

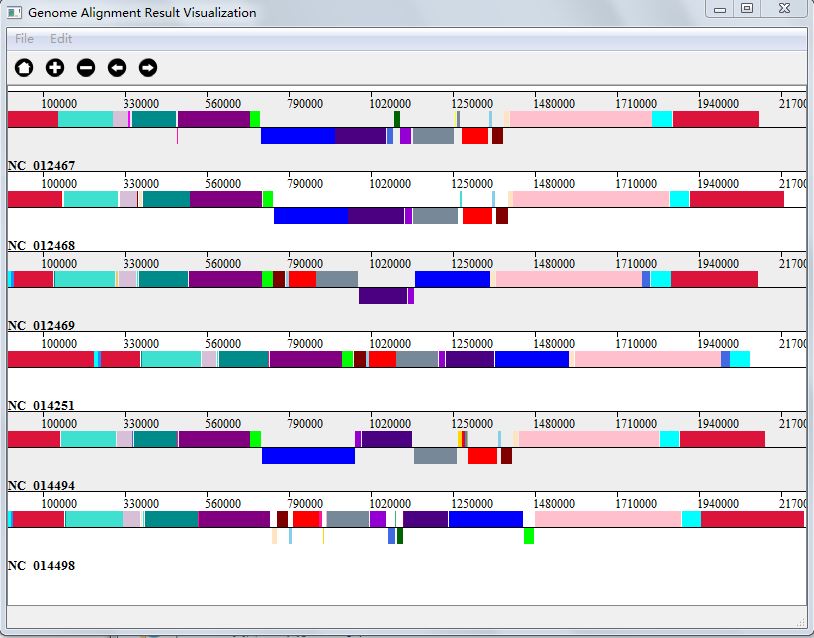
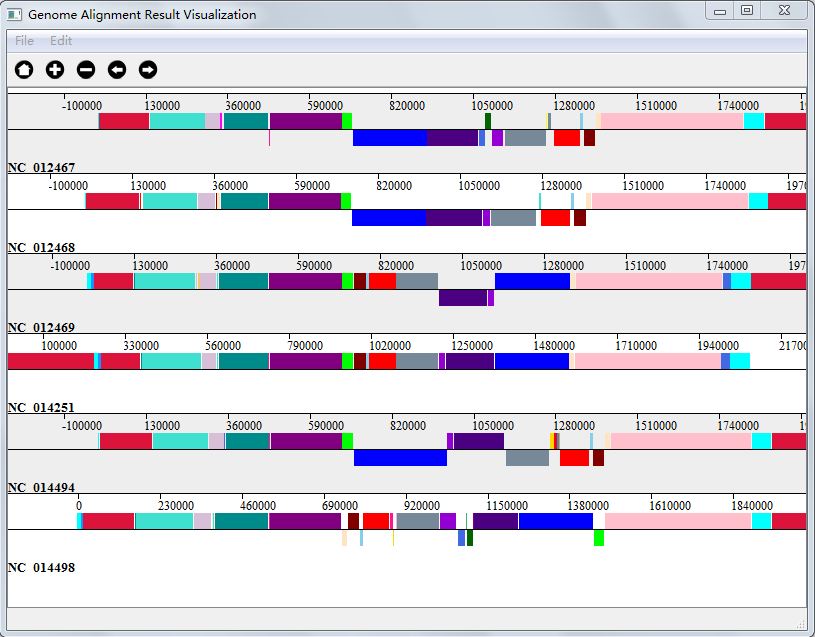
Genome alignment result is in the form of high conserved blocks in genomes, during visualization,we get the information of each block from alignment result, and then according to the site of genome, we draw each block in genome, and different aligned block will be in different color. Each colored line stand for a single genome, and one colored segment stands for one block with one color.
You can get your visualization result in two ways: either choose "Ok" when finishing alignment, or click "Genome Alignment Result Visualization" button in Genome Alignment interface(Figure 4.0.1). And Figure 4.4.1 is the visualization result of my genome alignment data.
You can save your visualization into JPG image file, or SVG file, by clicking the menu part of "File".
We develop many user interacting functions in out tool.
The first interacting function is you can choose which genome to display, and set genome order, by clicking the menu part of "Edit"-->"Genome List". Figure 4.5.1 shows the interface window that can change genomes and order.
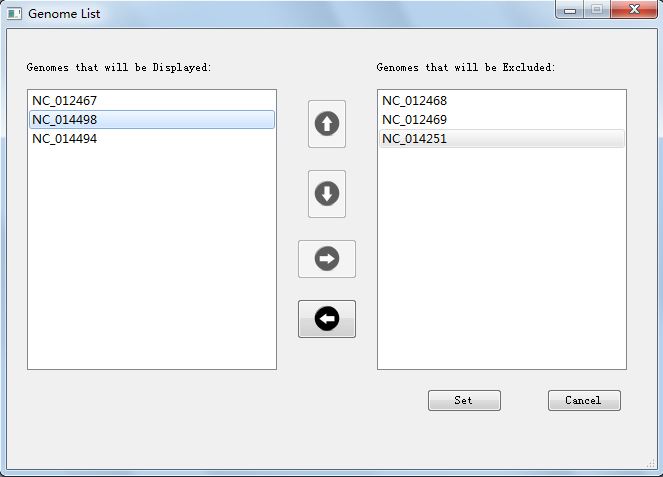
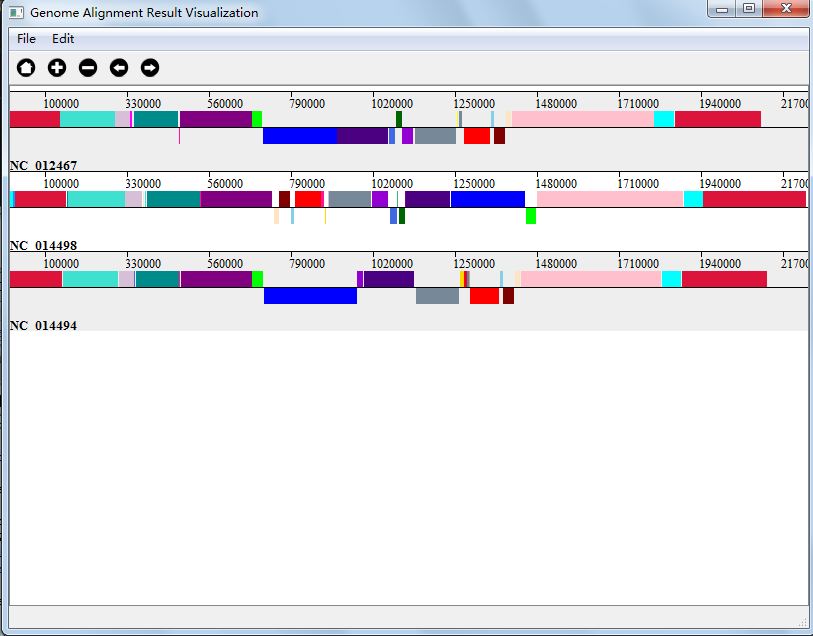
Here we just choose three genomes and change their order:NC_012467, NC_014498 and NC_014494. We can see the result in Figure 4.5.2.
The second interacting function is that you can align the genome by nucleotide site, by double clicking a site with your mouse. In Figure 4.5.3 , we double click a purple site in genome NC_014251.
The third interacting function is toolbar, you can zoom in, zoom out, move right, move left, and go back to the initial state.
 Go to initial state
Go to initial state Zoom in
Zoom in Zoom out
Zoom out Move left
Move left Move right
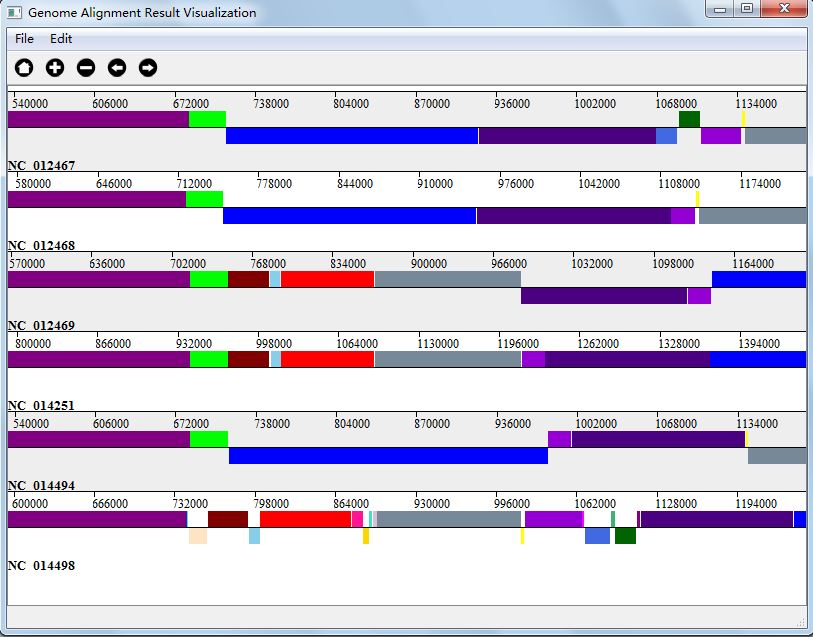
Move rightFigure 4.5.4 shows a visualization result of zoom in.
<< 4.2 4.3 |
5.Get Orthologs Clusters and Visualization Result >> |
|