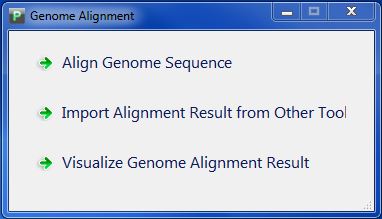
Click button "Structure Analysis" in PGAP-X Lite Launcher, you will get the "Structure Analysis" interface like Figure 4.0.1. Including three functional buttons:
Click button "Align Genome Sequence", you will enter the genome alignment interface like Figure 4.1.1.
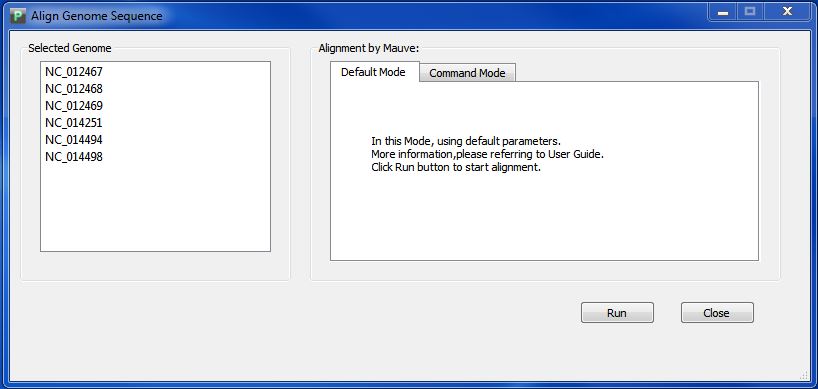
In default mode ,we will call MAUVE in background. you just need a single click on button "Run".
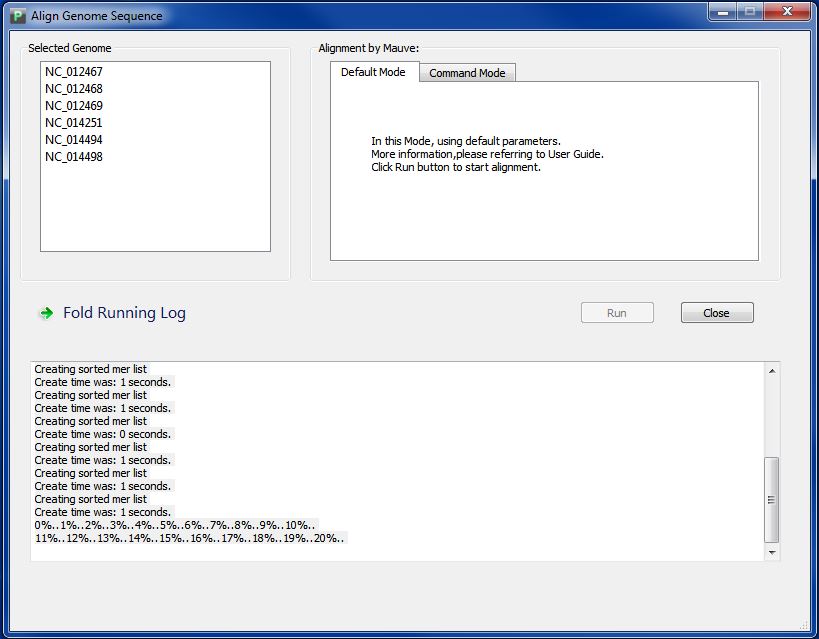
Wait for seconds, and you will see the interface like Figure 4.1.2, you can see the aligning process in the buttom part. It may takes you minutes to hours for alignment, depending on the genome number and length.
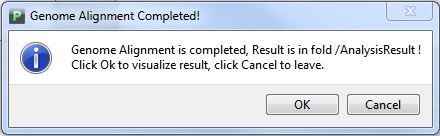
When finished, you will get a message like Figure 4.1.3, Click button "Ok", you will get your alignment visualization result directly. At the same time, you can find your alignment result in the folder "GenomeAlignmentResult" in your workspace, and there are two files in the folder,which are "WholeGenomeAlignment.xmfa", and "GenomeName.list". Do NOT try to change its name.
<< 3.Get Start |
4.2. 4.3 >> |
|