Click button "Orthologs Analysis" in PGAP-X Lite Launcher, you will get the "Orthologs Analysis" interface like Figure 5.0.1. Including three functional buttons:
| You are here: PGAP-X Home > Manual > Get Orthologs Clusters and Visualization Result | ||||
Click button "Orthologs Analysis" in PGAP-X Lite Launcher, you will get the "Orthologs Analysis" interface like Figure 5.0.1. Including three functional buttons:

Click button "Identify Orthologs Clusters", and you will get the interface of ortholog analysis like Figure 5.1.1.
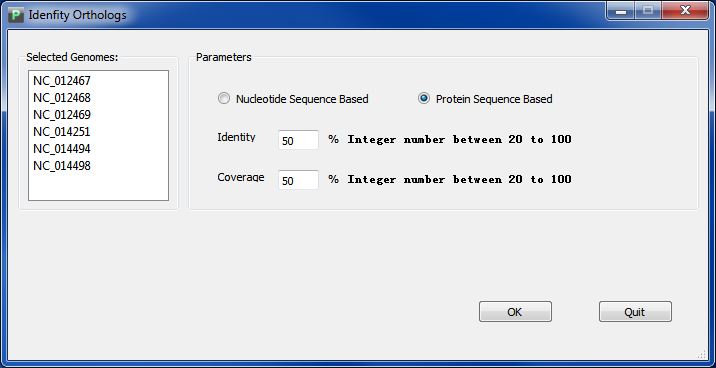
You can choose to analyze orthologs clusters using genome sequence or protein sequence, enter your parameter and click button "Ok", and the analyzing process will start. When process finished, there will automaticly a window presenting the visualization result of orthologs clusters. You can also find your ortholgs clusters data in folder "GenomeOrthologsCluster" in your workspace, the file name will be "OrthologsCluster.reference", and do NOT try to change its name inside the workspace.
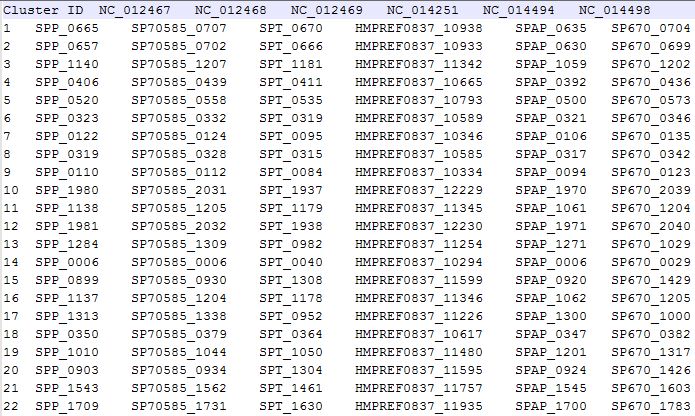
Figure 5.2.1 is a sample of OrthologsCluster.reference file.
Click button "Import Ortholog Clusters Results", you will enter the import interface like Figure 5.2.1, You can import your ortholog clusters file.
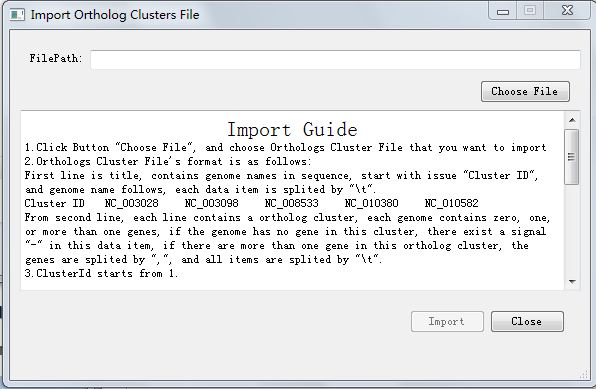
Click Button "Choose File", and choose Orthologs Cluster File that you want to import.
Orthologs Cluster File's format is as follows:
First line is title, contains genome names in sequence, start with issue "Cluster ID", and genome name follows, each data item is splited by "\t".
Cluster ID NC_012467 NC_012468 NC_012469 NC_014251 NC_014494 NC_014498
From second line, each line contains a ortholog cluster, each genome contains zero, one, or more than one genes, if the genome has no gene in this cluster, there exist a signal "-" in this data item, if there are more than one gene in this ortholog cluster, the genes are splited by ",", and all items are splited by "\t".
ClusterId starts from 1.
When finished, the button "Import" will be available. Click Import button and file you chosen will be import to your workspace, Pay attention: your former file will be overlaped.
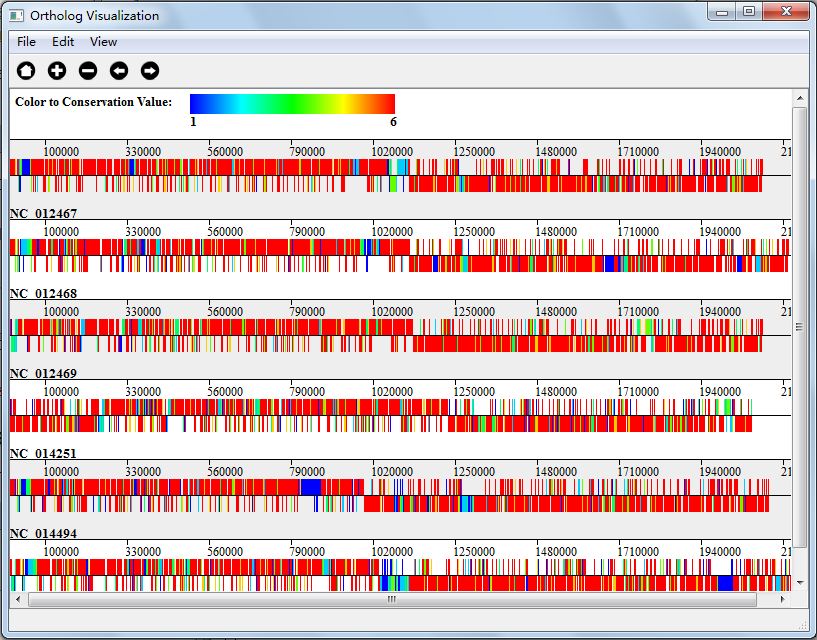
Ortholog analysis results are stored in the form of ortholog clustes, each cluster contains genes that may have the same ancestors, these genes may have the same functions. The number of genes in each cluster is called conservation value of the cluster. For instance, if the cluster have 4 genes, then the conservation of the cluster is 4, each gene in this cluster also have a conservation value of 4. We paint each gene on its genome according to its sites on genome, and we mark gene with different color according to its conservation value.
You can get your visualization result in two ways: either when finishing ortholog analysis, or click "Orthologs Analysis Result Visualization" button in Orthologs Analysis interface(Figure 5.0.1). And Figure 5.3.1 is the visualization result of my genome alignment data.
You can save your visualization into JPG image file, or SVG file, by clicking the menu part of "File".
<< 4.4 4.5 |
5.4 >> |
|